OCTSEG
What is OCTSEG?
OCTSEG (Optical Coherence Tomography Segmentation and Evaluation GUI) is a graphical user interface (GUI) written in MATLAB for research purpose. The retinal layers and the blood vessels of retinal OCT scans can be segmented. Tools for the manual correction of the automated segmentations are provided. The segmentations or resulting thickness measurements on the retinal layers may be exported as a CSV file, which is readable by many standard software programms (e.g. Excel).
The main reason for the development of the GUI was to enable our clinical research partners in the project  Advanced Fundus Imaging for Early Detection of Eye Diseases the use of our software algorithms in clinical studies.
Advanced Fundus Imaging for Early Detection of Eye Diseases the use of our software algorithms in clinical studies.
The source code of OCTSEG can be downloaded below.
Current Features
- Heidelberg Engineering Spectralis OCT RAW data (.vol ending): Circular scans and Optic Nerve Head centered volumes are supported
- Image file (.tif, .pgm, .jpg) support. Multiple images can be read in as volumes using user generated .list files (a text file format). Note: The algorithms have never been tested on other data then from the Spectralis system. If you will try it out: Please tell me about the results, I am always interested!
- Automated segmentation of 6 prominent retinal layers (including the inner limiting membrane, outer nerve fiber layer boundary, and retinal pigment epithelium)
- Automated segmentation of the blood vessel positions on circular scans
- Batch processing of circular scans
- Manual correction of possible segmentation errors
- Visualization of the data and the segmentation results, including enface views and thickness maps
- Export of the segmentation results to CSV text files
Screenshots
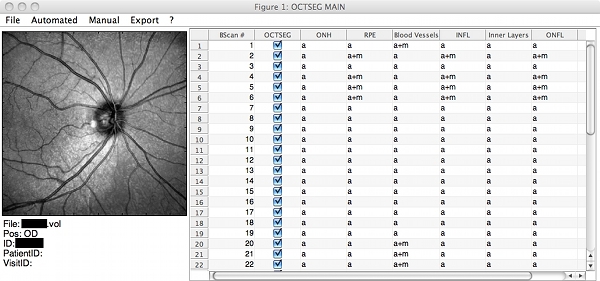 |
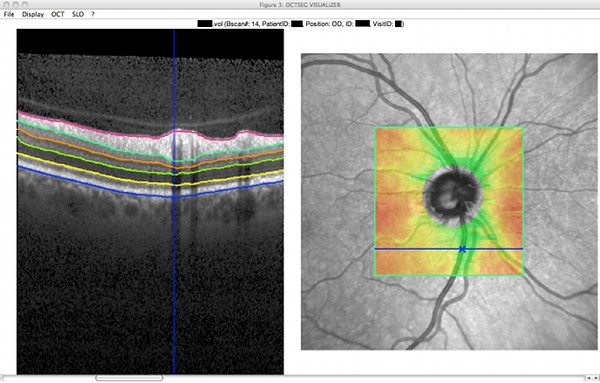 |
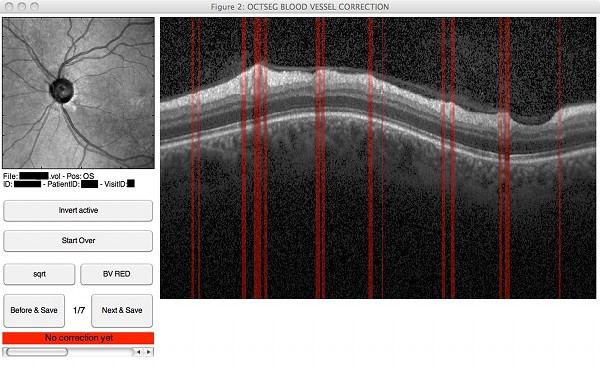 |
 |
Version information
The current and final version is version v0.4 (published only as source code).
v0.4 (January 2016)
- Revised the sources and source comments for source code publication.
- Added example B-Scans.
v0.35 (August 2012)
- Major Bugfix: Thickness exports of layers other than RNFL and retina only exported automated segmentation results, even if a manual segmentation was available. Fixed.
v0.34 (July 2012)
- Color indexed BMP files may now be opened.
- Minor bugfixes and compatibility improvements.
v0.33 (May 2012)
- Visualization of rotated volume scan patterns (e.g. the posterior pole scan pattern from Heidelberg engineering).
- Binary .meta file writeout (faster than text .meta files).
- Minor bugfixes.
v0.32 (December 2011)
- Support of image data (.tif, .jpg., .pgm) files and .list volumes enabled.
- Log intensity scaling added in the visualization tool.
- Subvolume export enabled.
v0.31 (July 2011)
- Additional CSV data export options: All layer group thicknesses.
v0.3 (July 2011)
- Minor Bugfixes/ Parameters changes
- New entry in octsegMain: "Optimization". Preparation to enable better volume segmentations.
v0.23 (June 2011): Minor Bugfix
v0.22 (June 2011)
- Major change: Internal processing changes: More data is reused, slight speed increase
- Minor change: Blood vessel segmentation is now a mandatory precondition for the inner layer segmentation.
- Minor change: VISU Slo menu restructured, visualization of blood vessel positions on volume scans
v0.21 (May 2011): Minor bugfix.
v0.2 (May 2011): Major changes:
- Internal changes resulting in a speed gain and less memory usage.
- Zooming and scaling possibilities in the visualizer, segmentation correction and blood vessel position correction.
v0.11 (May 2011): Minor bugfixes:
- Blank signs in the data path are now supported
- A complete fail of the ONH segmentation does not prevent further segmentations anymore.
v0.1 (May 2011): First preliminary version
Important (1): This software is research software. Expect no perfect stability and user friendlyness. Only a very rudimentary manual is provided. No warranty or any other guaranties are given. Use the software only for research purposes and at your own risk!
Important (2): The development of OCTSEG has been discontinued. Please understand that no further support can be given - but you may help yourself and use the sources to continue working on the program or just use the parts you find helpful for your own research!
Download
The use of the software is free, but you may want to search our  publications for references that are worth citing in your next article. An installer for the compiled version for Windows including the Matlab Runtime Environment can be downloaded here:
publications for references that are worth citing in your next article. An installer for the compiled version for Windows including the Matlab Runtime Environment can be downloaded here:
![]() octsegInstaller.exe (v0.35, 181 MB)
octsegInstaller.exe (v0.35, 181 MB)
If you already have the appropriate Matlab Runtime installed (i.e. you installed OCTSEG once using the package above), you only have to replace the .exe and .param files. These are provided, together with the preliminary manual, in the following package:
![]() octseg.zip (v0.35, 2.2 MB)
octseg.zip (v0.35, 2.2 MB)
The OCTSEG sources (Matlab code) including example B-Scans can be downloaded here:
![]() octsegSource.zip (v0.4, 5.1 MB)
octsegSource.zip (v0.4, 5.1 MB)
A short preliminary manual is included in the package. Read it before starting to work with the programm.
Authors & Contributors
The OCTSEG software is developed by  Markus Mayer as main author.
Markus Mayer as main author.
Kris Sheets (Retinal Cell Biology Lab, Neuroscience Center of Excellence, LSU Health Sciences Center, New Orleans, LA) supported the development with providing bugfixes and suggestions for improvements. You can find his ImageJ plugin for opening Heidelberg Engineering Spectralis RAW data files here.


 +49-9131-85-27775
+49-9131-85-27775
 +49-9131-85-27270
+49-9131-85-27270
