
Simone Gaffling M. Sc.
Alumnus of the Pattern Recognition Lab of the Friedrich-Alexander-Universität Erlangen-Nürnberg
Landmark-constrained 3-D Histological Imaging: A Morphology-preserving Approach
Landmark-constrained 3-D Histological Imaging: A Morphology-preserving Approach
Motivation
To achieve smooth 3-D reconstruction of histological image sequences, an unwarping strategy is required to reverse tissue deformations due to slicing. This is done using non-rigid registration. The strategy how the registration is applied to the slice sequence, however, plays an important role. Applied straight forward - using one image as reference, and matching the others against it - can lead to the so-called banana-effect (see below), and other unwanted behavior.
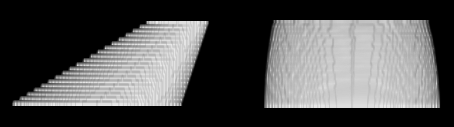
Figure on left: Reconstructions using synthetic
images. Left: Stacked original slices. Right: Non-rigid registration mostly optimizes overlap and smooth contour progression, but changes original object.
To prevent this effect, we propose to first generate an approximation of the morphology using trajectories. These trajectories are generated by tracking prominent anatomical features along the slice sequence, and approximating these points by a smooth function. The offset of the originally selected point to the calculated trajectory is then used as ground-truth deformation that is applied during non-rigid registration.
Method
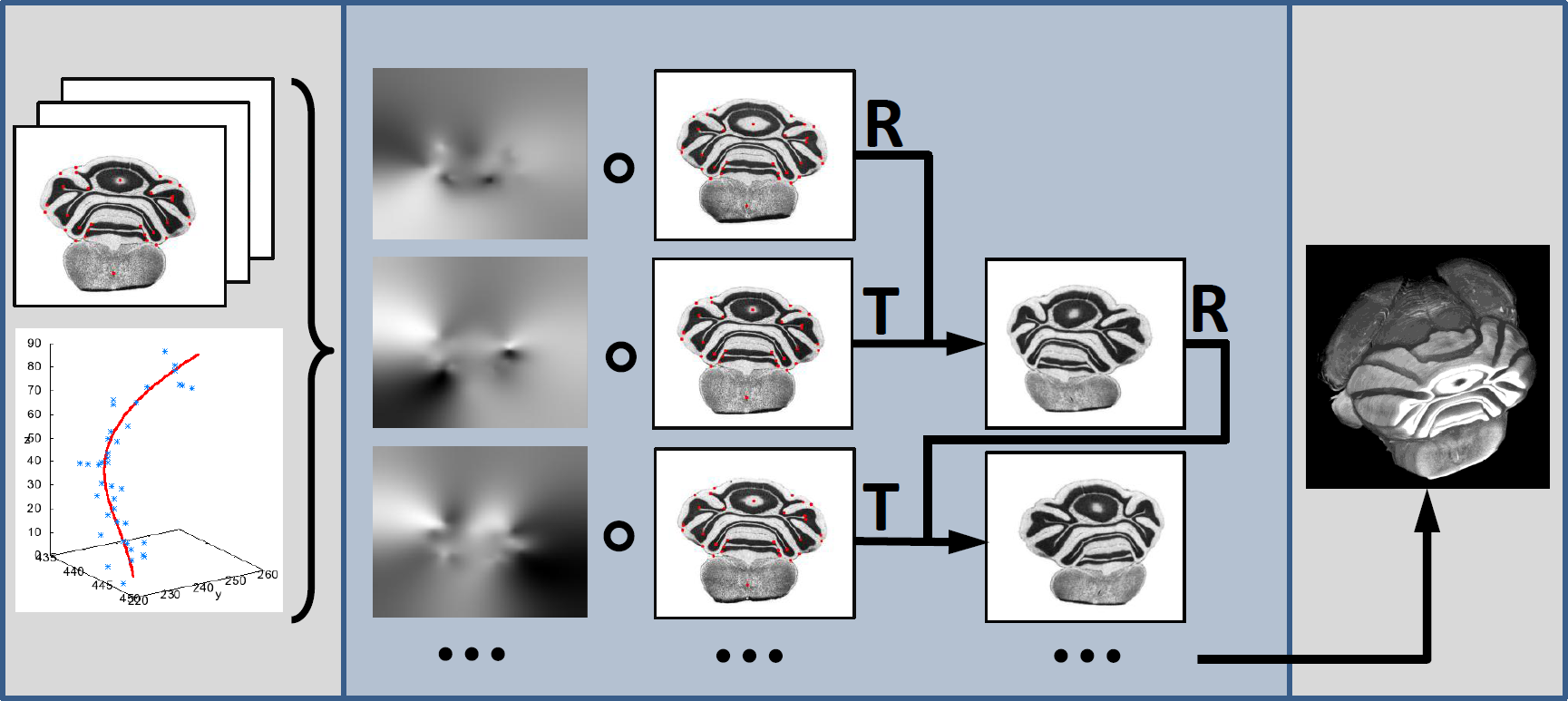
First, prominent anatomical features are manually selected in the slice sequence. The trajectories are created using an order 4 polynomial fitting procedure.
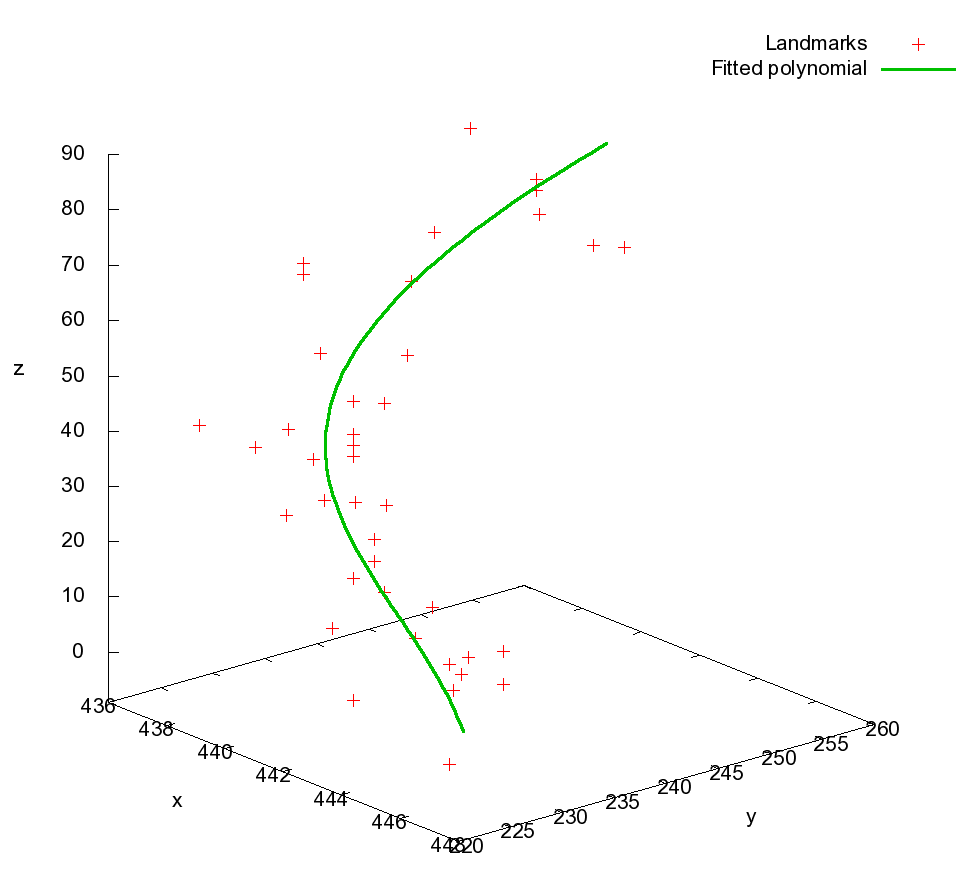
Left: Landmarks belonging to the same morphological structure, and their fitted trajectory.
The offset from an originally selected point to the intersection of the trajectory with the respective slice is the estimated true location the respective landmark should ideally lie on. All landmark offsets of one slice are then transformed into an initial deformation field. The known transformations are used as Dirichlet boundary conditions for the computation of the final deformation field, e.g., they are kept constant over the iterations. The neighborhood of these points is influenced by the curvature-regularizer used in the non-rigid registration procedure.
Below you can see the final employed workflow for our proposed reconstruction scheme.
Results
We tested our approach on two data sets. First, we created a synthetic image sequence comprised of a white disk with changing diameter and position representing a diagonal structure, and two white rectangles with fixed position on black background.
The second data set is a histological data set consisting of the first 100 of 350 Nissl-stained cryo sections of an adult mouse brain, available online by ![]() Ju et al.
Ju et al.
The parameters of the non-rigid registration were set to prevent strong warps and remaind unchanged for all experiments. We compared our approach agains a merely rigidly registered reconstruction, and a reconstruction using non-registration between an image, and its already registered and deformed predecessor.
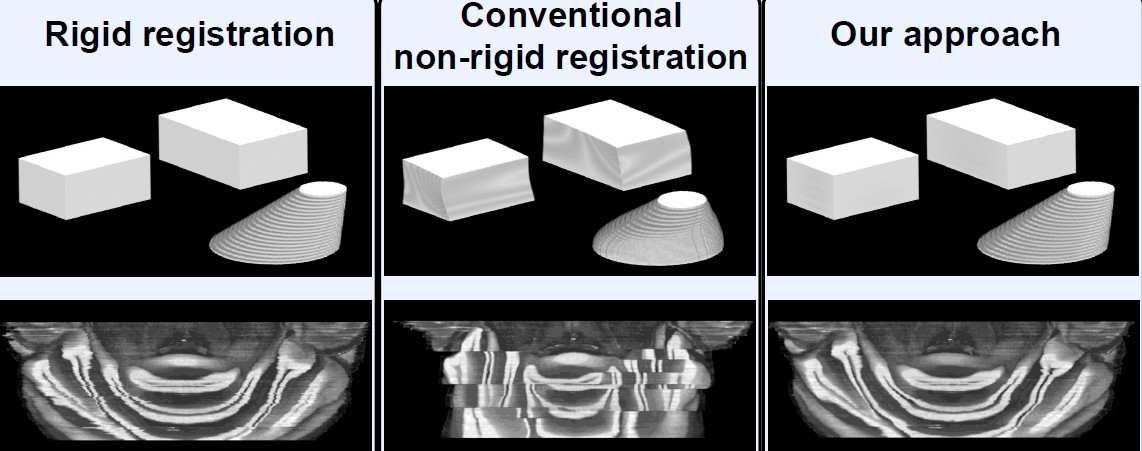
Top: Synthetic slices.
Bottom: Histological slices.
Left: Rigid registration and stacking shows jagged contours, originating from the slice deformations.
Middle: Without landmarks, the structures are matched very well, but the banana effect is clearly visible. Matching against the deformed preceding slice creates distinct blocks where the optimization routine got stuck in local minima.
Right: Relocating the landmarks on the morphology model preserves the curvature of brain structures. The novel registration routine guarantees exact positioning of the landmarks, while still smoothing contours.
The bibliography information for the article about this approach can be found ![]() here.
here.

 +49 9131 85 27874
+49 9131 85 27874
 +49 9131 85 27270
+49 9131 85 27270

