
Simone Gaffling M. Sc.
Alumnus of the Pattern Recognition Lab of the Friedrich-Alexander-Universität Erlangen-Nürnberg
Retrospective Microscopy
Nowadays, the three-dimensional (3D) visualization of scientific data is commonly used in a broad range of research areas to gain more insight in different processes. Biologists use it to visualize protein molecules to find new types of drugs, using the information of the 3D structure. In medical image processing it is also frequently used, especially to visualize anatomical and functional data from imaging devices like CT, MRI or PET/SPECT, to alleviate medical diagnoses.
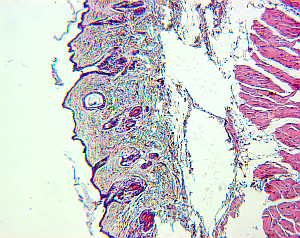
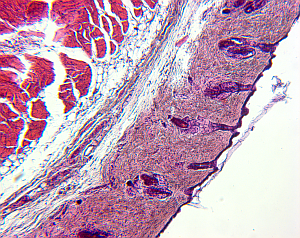
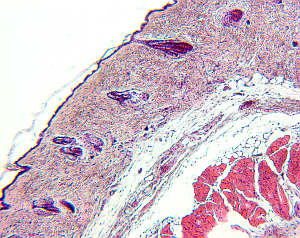
For small structures like cells, it is a common approach to create high-resolution microscopic images of the structures of interest. To do so, the tissue is cut in slices and visualized in the microscope, and based on the two-dimensional images the original 3D structure is envisioned. However, this process is hard, leaving room for interpretation, and in addition, it is highly subjective depending on the person viewing the data.
 |  |  |
|---|
Therefore, it would be beneficial to reconstruct the original 3-D structure from the set of histological images, to alleviate the investigation of structure properties like morphological features, and to remove the factor of subjectivity.
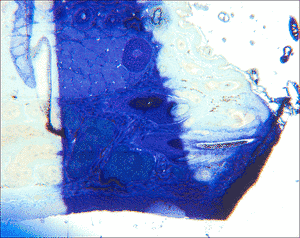
For this task, a software, named MicroSlice Visualization, was developed at the Chair of Pattern Recognition, University of Erlangen-Nuremberg. It is used to process a data set of histological images with the goal of reconstructing the 3D structure. Those data sets are obtained by taking photographs of the tissue viewed under a microscope. Due to the mechanical processes during the preparation steps, the final images show a multitude of artifacts. Common are differences in intensities, blurring, and disrupted slices, as well as deformations and post-mortal artifacts like holes.
MicroSlice Visualization is able to perform the basic steps necessary for a 3-D reconstruction. Faulty slices can be labeled and excluded for further steps, and intensity inhomogeneities due to the staining are corrected. Then, a rigid registration of adjacent slices is performed to match their structures. A segmentation step enables to extract regions that are of special interest for the user, and finally the data set is resampled to build the 3D volume, that can later be visualized using a rendering software.
For more detailed information, see my project pages on
 Interpolation of Histological Images
Interpolation of Histological Images 3-D Histomorphometric Reconstruction
3-D Histomorphometric Reconstruction Sorting of histological image sequences
Sorting of histological image sequences Landmark-constrained 3-D Histological Imaging
Landmark-constrained 3-D Histological Imaging

 +49 9131 85 27874
+49 9131 85 27874
 +49 9131 85 27270
+49 9131 85 27270

