Dipl.-Inf. Andreas Wimmer
Alumnus of the Pattern Recognition Lab of the Friedrich-Alexander-Universität Erlangen-Nürnberg
Semi-automatic Organ Segmentation
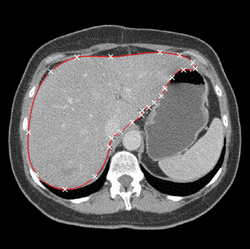
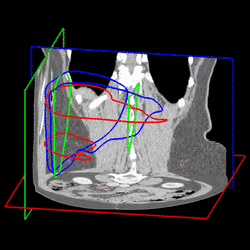
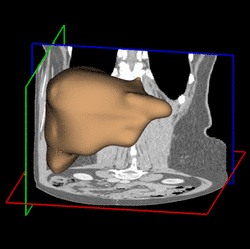
Various approaches have been proposed for the segmentation of organ shapes. Due to the limited soft-tissue contrast in computed tomography (CT) acquisitions, methods without shape knowledge often erroneously leak into other tissues with similar intensity values. In addition, pathologic structures like tumors with significantly different intensities are often missing in the segmentation, although they are considered to be part of the organ. Shape-based approaches have been proposed to ensure a robust segmentation. However, creating a shape model for an organ is a complex task and requires reference segmentations. Furthermore, a limited training set may lead to a model which is too restrictive and fails to capture variations often encountered for organs like the liver. In order to address these issues, we propose a semi-automatic segmentation algorithm. The idea is to have the user perform a manual delineation of the organ cross-section on a few slices. Such delineation can be easily performed using a spline curve, as shown in Fig. 1 (a). For smooth organ shapes, only a small number of control points is typically required. Manual delineations performed on six slices from three orthogonal views are shown in Fig. 1 (b). Radial basis functions are then employed to fit a smooth surface [1] through the contours, as illustrated in Fig. 1 (c).
| ||
(a) |
(b) |
(c) |
Fig. 1. (a) Manual delineation of the organ cross-section using a spline curve. Control points are shown as white crosses. (b) Contours drawn for six slices on three orthogonal views. (c) Surface reconstruction from the contours using radial basis functions. | ||
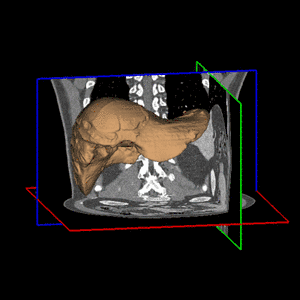
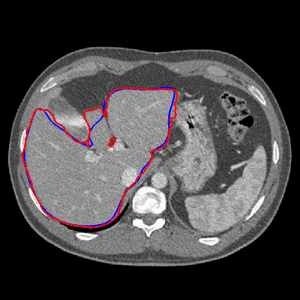
| In order to refine the segmentation, the initial surface is subsequently deformed by taking image information into account. Since the manual delineations provide valuable information about the true organ boundary, they are taken into consideration, too. In regions away from contours, image content prevails, whereas in regions close to contours, more weight is put on the shape information derived from the contours. Thereby, a shape model tailored to the specific data set is obtained. The paradigm is to perform delineations in regions where image information is ambiguous, e.g. at the border between liver tissue and muscle tissue. In other regions with higher contrast, image information prevails, relieving the physician from performing much delineation. The final surface reconstruction and the effect of the automatic refinement is shown in Fig. 2 for a different data set.
| ||
(a) |
(b) |
| Fig. 2. Final segmentation (a) and result of the automatic refinement for a slice (b). | |
[1] Carr, J.C., Beatson, R.K., Cherrie, J.B., Mitchell, T.J., Fright, W.R., McCallum, B.C., Evans, T.R.: Reconstruction and representation of 3D objects with radial basis functions. In: SIGGRAPH ’01: Proceedings of the 28th annual conference on Computer graphics and interactive techniques. (2001) 67–76 | |