
Simone Gaffling M. Sc.
Alumnus of the Pattern Recognition Lab of the Friedrich-Alexander-Universität Erlangen-Nürnberg
Sorting of Histological image sequences
Motivation
The 3-D reconstruction of histological image sequences relies heavily on the consistency of the data sets, especially artifact-free images and their proper order. However, since the preparation of the data sets is done manually, in particular the slicing and digitizing, it is very probable that the final image sequence shows disconitinuities, which then prevent a proper reconstruction. Therefore a sequence consistency check should be performed, and - if found necessary - the proper order should be restored.
Methods
Since it is not possible to define a "<" or ">" relation on images easily, normal sorting algorithms are not applicable to solve this problem. Instead, we use the - in most cases - valid assumption that neighboring or close images should be more similar to each other than more distant slices.
Having an image sequence, the distance is calculated between one slice to the next using rigid registration of the images and calculation of the Mean Squared Distance (MSD). Then, a threshold is derived using the mean distance and standard deviation of the distances, and outliers are detected. The sequence is divided into consistent blocks regarding this threshold.
Then, the similarity between all block borders is calculated, and the blocks are matched and rearranged according the best found similarities. The reordered sequenced is then used as input for the next iteration. The algorithm stops after a maximum number of iterations, or if no changes are found.
The approach was tested on a histological data set of a mouse brain, which was specially prepared for reconstruction, and can be considered mostly artifact free.
Results
The method is able to find discontinuities in image sequences, and to restore the sequence in most cases. The runtime depends for the greater part on the speed of the rigid registration method, and the degree of parallelization.
Problems, however, arise due to image artifacts like tissue deformations and missing images. Futhermore, the choice of the threshold for outlier detection is too unflexible to handle real datasets, which demands further investigation and refinement.
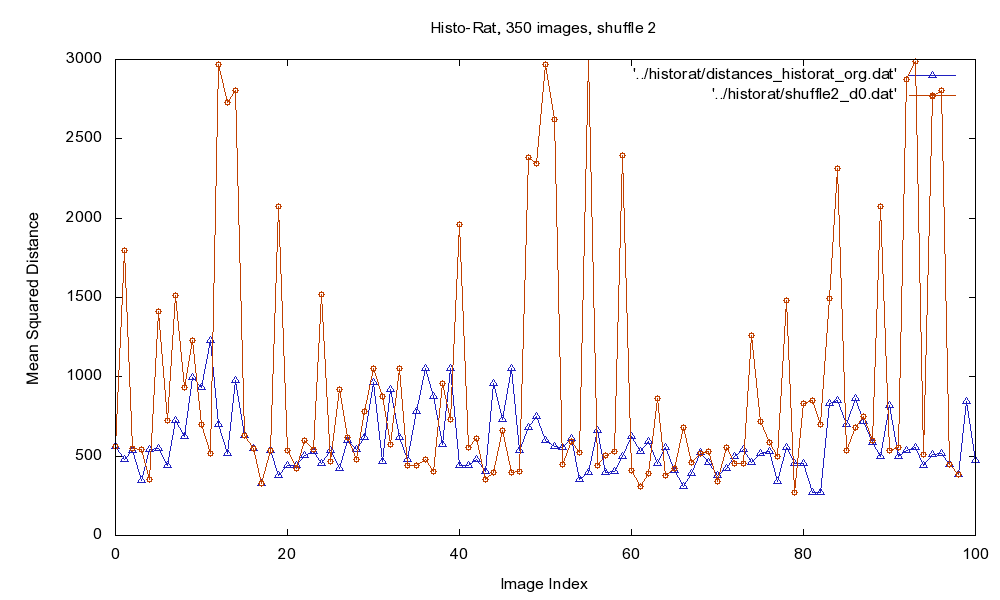
Fig.1: Mean Squared Distance (MSD) of original image sequence (blue), and for the intentionally shuffled sequence (red).
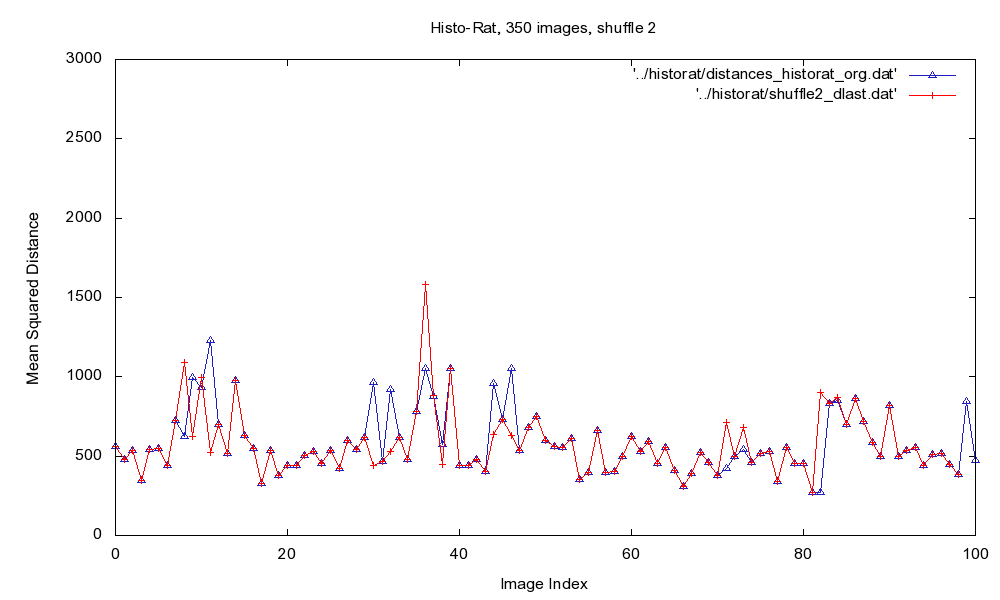
Fig.2: Plot of MSD of the original image sequence (blue), and after the final iteration of the sorting algorithm (red).
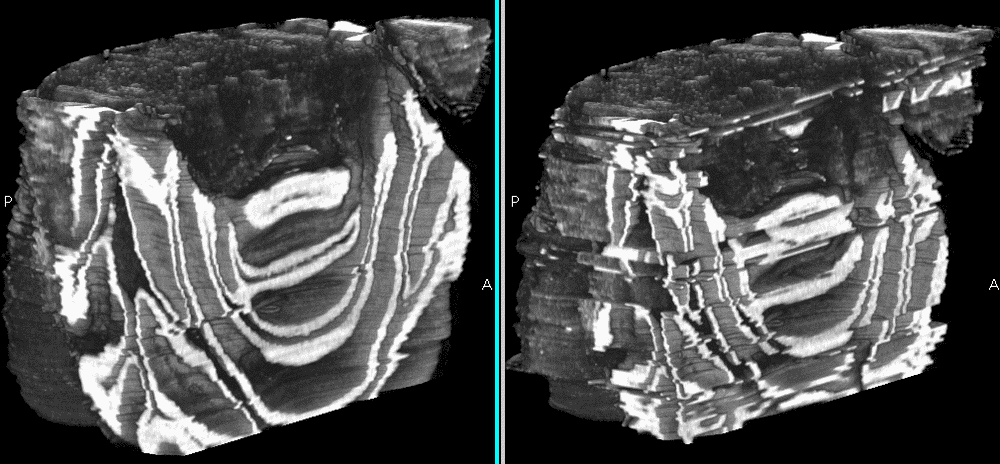
Fig.3: Reconstruction images. Right: Shuffled data set. Left: Sorted.

 +49 9131 85 27874
+49 9131 85 27874
 +49 9131 85 27270
+49 9131 85 27270

