
Friedrich-Alexander-Universität Erlangen
Lehrstuhl für Mustererkennung
Martensstraße 3
91058 Erlangen

We aim to improve current cell image processing by modern approaches, such as deep learning-based segmentation and classification, as well as image processing to improve the quality of said images.
We target on modalities such as digital histology whole slide images (see image on right side) and confocal laser endomicroscopy images.
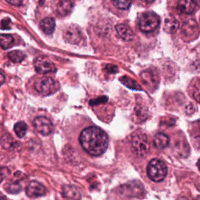
One modality we target is digital histology images (see image on right side). The image size produced by todays slide scanners is large (up to 90k times 90k pixels), and the number of singular cells on the images is thus high.
We aim to help pathologist in a faster and more accurate diagnosis by selecting diagnostically relevant structures in WSI, or by automatically classifying all relevant cells - a task that would be too tedious for a human expert. This project is done in close cooperation with the working group of Prof. Robert Klopfleisch (Institute for Veterinary Pathology, Freie Universität Berlin).
We believe that training and evaluation of such algorithms is heavily dependent on the availability of high quality expert-annotated data sets. To facilitate work in this area, we want to encourage other research groups to work on the data sets provided by us (soon to be published).
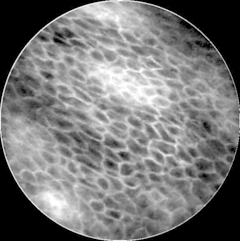

Confocal Laser Endomicroscopy (CLE) is a fiber-bundle based in-vivo, in-situ imaging method, enabling non-invasive micro-structural analysis of tissue. CLE was proven to be valuable in many anatomic locations, including the upper respiratory tract, the gastro-intestinal tract and the brain.
The goal of this project is to detect cancerous tissue in CLE images of the oral cavity and the vocal cords. The current treatment of these diseases is a histological analysis of specimen and a surgical resection, which has a rather high long-term survival rate, or radiation therapy with a lower survival rate. An early detection of cancerous tissue could lead to a lowered complication rate for further treatment, as well as a better overall prognosis for patients. Further, an in-vivo diagnosis during operation could narrow down the area for the necessary surgical excision, which is especially beneficial for cancer of the vocal cords.
Besides classification of images into tumor and normal tissue, we also investigated detection of common artifacts in CLE images.