Dr.-Ing. Andreas Fieselmann
Alumnus of the Pattern Recognition Lab of the Friedrich-Alexander-Universität Erlangen-Nürnberg
DIPPO - A Digital Brain Perfusion Phantom
1. Overview
Brain perfusion imaging with C-arm CT is currently challenged by the slow C-arm rotation (see project page). In order to evaluate novel dynamic image reconstruction algorithms and scanning protocols a digital brain perfusion phantom has been designed.
On this website, the phantom is described and relevant software implementations are provided in order to improve the reproducability of our results that are presented in [1].
2. Phantom Design: Static Phantom Components
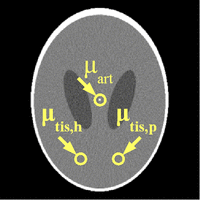
We propose to use a Shepp-Logan-type phantom [2,3] to model the static components of the brain (skull, ventricles, non-perfused brain tissue). Our phantom (Figure 1) has been slightly adapted with respect to the original Shepp-Logan phantom [2]. For example, the two ventricles are symmetrical in order to avoid any influence of spatial unsymmetries. Note, the main purpose of our phantom is to provide realistic temporal dynamics of contrast agent flow.
Kak and Slaney [4] provide a description how a Shepp-Logan-type phantom can be constructed from a superposition of different ellipsoids. The parameters of the ellipsoids, cf. [4], of the Shepp-Logan-type phantom that we use are shown in Table 1. The phantom contains three regions of interest (ROI) (ellipsoids 5,6,7) with dynamically changing attenuation values that model an artery, a healthy tissue region, and a pathological tissue region, respectively.
The simulated artery is located in the center of the phantom in order to improve the quantification of reconstruction artifacts around the artery. If the artery was located off-center then the reconstruction artifacts would depend more severly on the trajectory (start and end view angle) of the X-ray source.
| ellipsoid | description | center [mm] | axis lengths [mm] | rotation angle [°] | X-ray attenuation [μ_w] |
| 1 | skull | (0.00, 0.00) | (92.00, 69.00) | 90 | 2 |
| 2 | brain tissue | (0.00, -1.84) | (87.40, 66.24) | 90 | -1 |
| 3 | left ventricle | (22.00, 0.00) | (31.00, 11.00) | 72 | -0.05 |
| 4 | right ventricle | (-22.00, 0.00) | (31.00, 11.00) | 108 | -0.05 |
| 5 | artery | (0.00, 0.00) | (1.00, 1.00) | 0 | dynamic |
| 6 | healthy tissue | (20.00, -60.00) | (2.00, 2.00) | 0 | dynamic |
| 7 | pathological tissue | (-20.00, -60.00) | (2.00, 2.00) | 0 | dynamic |
Table 1. Parameters of the ellipsoids of the phantom (see Figure 1). The X-ray attenuation is given relative to the X-ray attenuation μ_w of water.
3. Phantom Design: Dynamic Phantom Components
In this section, the dynamic change of the attenuation values in the artery and the two tissue regions will be briefly described. Note, a detailed mathematical description how the time-attenuation curves are created is provided in [1].
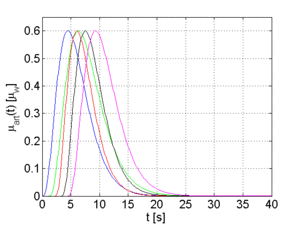
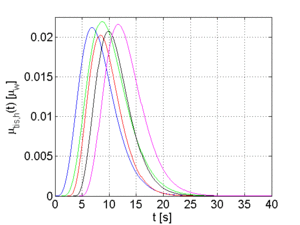
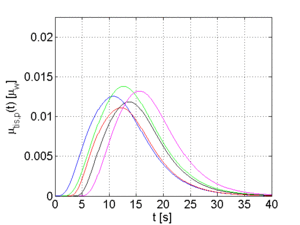
The arterial time-attenuation curve μ_aif(t) is modeled by a gamma-variate function. A discussion why a gamma-variate function can be applied to model contrast agent flow is provided in [5]. The tissue time-attenuation curves are computed from the arterial curve using the indicator-dilution theory [6]. Two different tissue curves are computed for each arterial curve which are representative for healthy (μ_trf,h(t)) and pathological (μ_trf,p(t)) tissue, respectively.
The full width at half maximum (FWHM) and the temporal offset of the arterial function can be varied in order to generate different sets of time-attenuation curves. Examples for 5 sets of curves, each set has a certain FWHM and temporal offset for μ_aif(t), are shown in Figure 2.
The following MATLAB functions (tested using MATLAB R2009a) generate the time attenuation curves for the artery and the two tissue regions. The "_example" file will generate the time curves that are actually shown in Figure 2. Please refer to the documentation in the files for further information how to use these files.
3. Evaluation Strategy
There are various perfusion parameters such as cerebral blood flow (CBF), cerebral blood volume (CBV), mean transit time (MTT) [6]. The most frequently employed method to compute these parameters uses a deconvolution of the arterial and the tissue time-attenuation curves. Since the measured data is corrupted by noise the deconvolution is in fact a discrete ill-posed problem which requires regularization.
The following MATLAB functions use the truncated singular value decomposition (TSVD) algorithm which is commonly applied in perfusion image analysis. Please refer to the documentation in the files for further information how to use these files.
4. References
- A. Fieselmann, A. Ganguly, Y. Deuerling-Zheng, M. Zellerhoff, C. Rohkohl, J. Boese, J. Hornegger, and R. Fahrig. Interventional 4-D C-Arm CT Perfusion Imaging Using Interleaved Scanning and Partial Reconstruction Interpolation. IEEE Transactions on Medical Imaging, vol. 31, no. 4, pages 892-906, 2012 (pdf, link)
- L. A. Shepp and B. F. Logan. The Fourier Reconstruction of a Head Section. IEEE Transactions on Nuclear Science, vol. 21, no. 3, pages 21-43, 1974. (pdf)
- P. Toft. The Radon Transform - Theory and Implementation. Ph.D. thesis. Department of Mathematical Modelling, Technical University of Denmark, 326 pages, 1996. (pdf: Part III - Appendices)
- A. C. Kak and M. Slaney. Principles of Computerized Tomographic Imaging. IEEE, Bellingham, WA, 1988. (pdf: Chapter 3)
- M. Mischi, J. A. den Boer, and H. H. M. Korsten. On the physical and stochastic representation of an indicator dilution curve as a gamma variate. Physiological Measurement, vol. 29, pages 281-294, 2008. (pdf)
- L. Østergaard, R. M. Weisskoff, D. A. Chesler, C. Gyldensted, and B. R. Rosen. High resolution measurement of cerebral blood flow using intravascular tracer bolus passages. part I: Mathematical approach and statistical analysis. Magnetic Resonance in Medicine, vol. 36, no. 5, pages 715-725, 1996. (abstract)